Why, would that change your answer?
I thought the main problem with evolution was that it predicts nothing and can't be disproven.
You thought wrongly. Or more likely, you made the mistake of believing the creationist propaganda on that topic.
If I'm wrong enlighten me. I'd like to try out some of the tests.
Here you go: 29+ Evidences for Macroevolution: The Scientific Case for Common Descent . Note that each section describes (in general) the kinds of predictions which evolutionary theory makes along each line of evidenciary research, and also explains what sorts of results would be potential falsifications (i.e. which kinds of results would count as failed predictions, and therefore a strong strike against the theory).
These of course are general overviews. For the actual vastness and thoroughness of the countless tests and verifications, and the mountains of confirming evidence, one would need to follow the links and citations on those pages, as well as peruse the relevant scientific journals and research archives.
For that matter, most of the workaday "meat and potatoes" research in biology actually are the result (and confirmation of) the predictions of evolutionary theory. For example, reconstruction of phylogenetic trees via comparative DNA analysis is only possible because of the predictions of evolutionary theory -- and if that theory was wrong, those DNA analyses would produce data which was incoherent garbage. However, the fact that such analyses invariably produce consistent, informationally coherent results, and even more to the point, produce results which *match* the results of independent studies of *other* molecular comparative studies *and* the data from the fossil and geologic record, is incredibly powerful confirmation of the many and varied types of specific predictions made by evolutionary theory.
For example, all of the following (from a previous post of mine), and the linked papers, are tests of, and successful confirmations of, the predictions of evolutionary theory:
Similar results (i.e., findings consistent with the predictions of evolutionary theory) are found when researching countless other lineages as well. Here's a similar overview of some of the findings of human/primate research, which again are confirmations of the predictions of evolutionary theory:Evolution of whales from terrestrial mammals
(From Plagiarized Errors and Molecular Genetics)That's just a quick layman-level overview of *one* of the many ways that whale evolution has been verified. For more technical examinations along several independent lines of evidence, see for example:.
A particularly impressive example of shared retroposons has recently been reported linking cetaceans (whales, dolphins and porpoises) to ruminants and hippopotamuses, and it is instructive to consider this example in some detail. Cetaceans are sea-living animals that bear important similarities to land-living mammals; in particular, the females have mammary glands and nurse their young. Scientists studying mammalian anatomy and physiology have demonstrated greatest similarities between cetaceans and the mammalian group known as artiodactyls (even-toed ungulates) including cows, sheep, camels and pigs. These observations have led to the evolutionist view that whales evolved from a four-legged artiodactyl ancestor that lived on land. Creationists have capitalized on the obvious differences between the familiar artiodactyls and whales, and have ridiculed the idea that whales could have had four-legged land-living ancestors. Creationists who claim that cetaceans did not arise from four-legged land mammals must ignore or somehow dismiss the fossil evidence of apparent whale ancestors looking exactly like one would predict for transitional species between land mammals and whales--with diminutive legs and with ear structures intermediate between those of modern artiodactyls and cetaceans (Nature 368:844,1994; Science 263: 210, 1994). (A discussion of fossil ancestral whale species with references may be found at http://www.talkorigins.org/faqs/faq-transitional/part2b.html#ceta) Creationists must also ignore or dismiss the evidence showing the great similarity between cetacean and artiodactyl gene sequences (Molecular Biology & Evolution 11:357, 1994; ibid 13: 954, 1996; Gatesy et al, Systematic Biology 48:6, 1999).
Recently retroposon evidence has solidified the evolutionary relationship between whales and artiodactyls. Shimamura et al. (Nature 388:666, 1997; Mol Biol Evol 16: 1046, 1999; see also Lum et al., Mol Biol Evol 17:1417, 2000; Nikaido and Okada, Mamm Genome 11:1123, 2000) studied SINE sequences that are highly reduplicated in the DNA of all cetacean species examined. These SINES were also found to be present in the DNA of ruminants (including cows and sheep) but not in DNA of camels and pigs or more distantly related mammals such as horse, elephant, cat, human or kangaroo. These SINES apparently originated in a specific branch of ancestral artiodactyls after this branch diverged from camels, pigs and other mammals, but before the divergence of the lines leading to modern cetaceans, hippopotamus and ruminants. (See Figure 5.) In support of this scenario, Shimamura et al. identified two specific insertions of these SINES in whale DNA (insertions B and C in Figure 5) and showed that in DNA of hippopotamus, cow and sheep these same two sites contained the SINES; but in camel and pig DNA the same sites were "empty" of insertions. More recently, hippopotamus has been identified as the closest living terrestrial relative of cetaceans since hippos and whales share retroposon insertions (illustrated by D and E in Figure 5) that are not found in any other artiodactyls (Nikaido et al, PNAS 96:10261, 1999). The close hippo-whale relationship is consistent with previously reported sequence similarity comparisons (Gatesy, Mol Biol Evol 14:537, 1997) and with recent fossil finds (Gingerich et al., Science 293:2239, 2001; Thewissen et al., Nature 413:277, 2001) that resolve earlier paleontological conflicts with the close whale-hippo relationship. (Some readers have wondered: if ruminants are more closely related to whales than to pigs and camels, why are ruminants anatomically more similar to pigs and camels than they are to whales? Apparently this results from the fact that ruminants, pigs and camels changed relatively little since their last common ancestor, while the cetacean lineage changed dramatically in adapting to an aquatic lifestyle, thereby obliterating many of the features -- like hooves, fur and hind legs -- that are shared between its close ruminant relatives and the more distantly related pigs and camels. This scenario illustrates the fact that the rapid evolutionary development of adaptations to a new niche can occur through key functional mutations, leaving the bulk of the DNA relatively unchanged. The particularly close relationship between whales and hippos is consistent with several shared adaptations to aquatic life, including use of underwater vocalizations for communication and the absence of hair and sebaceous glands.) Thus, retroposon evidence strongly supports the derivation of whales from a common ancestor of hippopotamus and ruminants, consistent with the evolutionary interpretation of fossils and overall DNA sequence similarities. Indeed, the logic of the evidence from shared SINEs is so powerful that SINEs may be the best available characters for deducing species relatedness (Shedlock and Okada, Bioessays 22:148, 2000), even if they are not perfect (Myamoto, Curr. Biology 9:R816, 1999).
Figure 5. Specific SINE insertions can act as "tracers" that illuminate phylogenetic relationships. This figure summarizes some of the data on SINEs found in living artiodactyls and shows how the shared insertions can be interpreted in relation to evolutionary branching. A specific SINE insertion event ("A" in the Figure) apparently occurred in a primitive common ancestor of pigs, ruminants, hippopotamus and cetaceans, since this insertion is present in these modern descendants of that common ancestor; but it is absent in camels, which split off from the other species before this SINE inserted. More recent insertions B and C are present only in ruminants, hippopotamus and cetaceans. Insertions D and E are shared only by hippopotamus and cetaceans, thereby identifying hippopotamus as the closest living relative of cetaceans (at least among the species examined in these studies). SINE insertions F and G occurred in the ruminant lineage after it diverged from the other species; and insertions H and I occurred after divergence of the cetacean lineage.
SINE Evolution, Missing Data, and the Origin of WhalesEvidence from Milk Casein Genes that Cetaceans are Close Relatives of Hippopotamid Artiodactyls
Analyses of mitochondrial genomes strongly support a hippopotamus±whale clade
A new Eocene archaeocete (Mammalia, Cetacea) from India and the time of origin of whales
Mysticete (Baleen Whale) Relationships Based upon the Sequence of the Common Cetacean DNA Satellite1
Eocene evolution of whale hearing
Novel Phylogeny of Whales Revisited but Not Revised
New Morphological Evidence for the Phylogeny of Artiodactyla, Cetacea, and Mesonychidae
Evolutionary theory predicted that there should be transitional forms between the reptilian-style jaw joint and the mammalian-style jaw joint (the earliest mammals evolved from reptiles). For years creationists crowed about the "missing links", and made their own predictions that not only would there be no such transitional fossils found, but that any creature with a jaw that was transitional between that of reptiles and mammals would die of starvation, since such a "half and half" jaw joint was "obviously" mechanically unworkable. Nonetheless, biologists kept searching for the fossils predicted by evolution, and not only found one or two, but found a *wealth* of them which provide a *very* complete and smooth transitional sequence -- exactly as evolution predicted. Oddly enough, I never hear the creationists bring that one up...Human/ape common ancestry:
This is just a taste of the massive amount of evidence for ape/human common ancestry, the amount for evolution in general (including between different specific animal families) would (and does) fill innumerable encyclopedias worth of volumes:
Or how about:Background: Retroviruses reproduce by entering a cell of a host (like, say, a human), then embedding their own viral DNA into the cell's own DNA, which has the effect of adding a "recipe" for manufacturing more viruses to the cell's "instruction book". The cell then follows those instructions because it has no reason (or way) to "mistrust" the DNA instructions it contains. So the virus has converted the cell into a virus factory, and the new viruses leave the cell, and go find more cells to infect, etc.
However, every once in a while a virus's invasion plans don't function exactly as they should, and the virus's DNA (or portions of it) gets embedded into the cell's DNA in a "broken" manner. It's stuck into there, becoming part of the cell's DNA, but it's unable to produce new viruses. So there it remains, causing no harm. If this happens in a regular body cell, it just remains there for life as a "fossil" of the past infection and goes to the grave with the individual it's stuck in. All of us almost certainly contain countless such relics of the past viral infections we've fought off.
However... By chance this sometimes happens to a special cell in the body, a gametocyte cell that's one of the ones responsible for making sperm in males and egg cells in females, and if so subsequent sperm/eggs produced by that cell will contain copies of the "fossil" virus, since now it's just a portion of the entire DNA package of the cell. And once in a blue moon such a sperm or egg is lucky enough to be one of the few which participate in fertilization and are used to produce a child -- who will now inherit copies of the "fossilized" viral DNA in every cell of his/her body, since all are copied from the DNA of the original modified sperm/egg.
So now the "fossilized" viral DNA sequence will be passed on to *their* children, and their children's children, and so on. Through a process called neutral genetic drift, given enough time (it happens faster in smaller populations than large) the "fossil" viral DNA will either be flushed out of the population eventually, *or* by luck of the draw end up in every member of the population X generations down the road. It all depends on a roll of the genetic dice.
Due to the hurdles, "fossil" retroviral DNA strings (known by the technical name of "endogenous retroviruses") don't end up ubiquitous in a species very often, but it provably *does* happen. In fact, the Human DNA project has identified literally *thousands* of such fossilized "relics" of long-ago ancestral infections in the human DNA.
And several features of these DNA relics can be used to demonstrate common descent, including their *location*. The reason is that retroviruses aren't picky about where their DNA gets inserted into the host DNA. Even in an infection in a *single* individual, each infected cell has the retroviral DNA inserted into different locations than any other cell. Because the host DNA is so enormous (billions of basepairs in humans, for example), the odds of any retroviral insertion event matching the insertion location of any other insertion event are astronomically low. The only plausible mechanism by which two individuals could have retroviral DNA inserted into exactly the same location in their respective DNAs is if they inherited copies of that DNA from the same source -- a common ancestor.
Thus, shared endogenous retroviruses between, say, ape and man is almost irrefutable evidence that they descended from a common ancestor. *Unless* you want to suggest that they were created separately, and then a virus they were both susceptible to infected both a man and an ape in EXACTLY the same location in their DNAs (the odds of such a match by luck are literally on the order of 1,000,000,000,000,000,000 to 1...), *and* that the infections both happened in their gametocyte cells (combined odds on the order of 1,000,000 to 1) *and* that the one particular affected gametocyte is the one which produces the egg or sperm which is destined to produce an offspring (*HUGE* odds against), and *then* the resulting modified genome of the offspring becomes "fixed" in each respective population (1 out of population_size^squared)...
Then repeat that for *each* shared endogenous retrovirus (there are many) you'd like to claim was acquired independently and *not* from a shared ancestor...
Finally, you'd have to explain why, for say species A, B, and C, the pattern of shared same-location retroviruses is always *nested*, never *overlapped*. For example, all three will share some retroviruses, then A and B will both share several more, but if so then B *never* shares one with C that A doesn't also have (or at least remnants of).
In your "shared infection due to genetic similarities" suggestion, even leaving aside the near statistical impossibility of the infections leaving genetic "scars" in *exactly* the same locations in independent infections, one would expect to find cases of three species X, Y, and Z, where the degree of similarity was such that Y was "between" X and Z on some similarity scale, causing the same disease to befall X and Y but not Z, and another disease to affect Y and Z but not X. And yet, we don't find this in genetic markers. The markers are found in nested sequence, which is precisely what we would expect to see in cases of inheritance from common ancestry.
Here, for example, is an ancestry tree showing the pattern of shared same-location endogenous retroviruses of type HERV-K among primates:
This is just a partial list for illustration purposes -- there are many more.
Each labeled arrow on the chart shows an ERV shared in common by all the branches to the right, and *not* the branches that are "left-and-down". This is the pattern that common descent would make. And common descent is the *only* plausible explanation for it. Furthermore, similar findings tie together larger mammal groups into successively larger "superfamilies" of creatures all descended from a common ancestor.
Any presumption of independent acquisition is literally astronomically unlikely. And "God chose to put broken relics of viral infections that never actually happened into our DNA and line them up only in patterns that would provide incredibly strong evidence of common descent which hadn't actually happened" just strains credulity (not to mention would raise troubling questions about God's motives for such a misleading act).
Once again, the evidence for common descent -- as opposed to any other conceivable alternative explanation -- is clear and overwhelming.
Wait, want more? Endogenous retroviruses are just *one* type of genetic "tag" that makes perfect sense evolutionary and *no* sense under any other scenario. In addition to ERV's, there are also similar arguments for the patterns across species of Protein functional redundancies, DNA coding redundancies, shared Processed pseudogenes, shared Transposons (including *several* independent varieties, such as SINEs and LINEs), shared redundant pseudogenes, etc. etc. Here, for example, is a small map of shared SINE events among various mammal groups:
Like ERV's, any scenario which suggests that these shared DNA features were acquired separately strains the laws of probability beyond the breaking point, but they make perfect sense from an evolutionary common-descent scenario. In the above data, it is clear that the only logical conclusion is that, for example, the cetaceans, hippos, and ruminants shared a common ancestor, in which SINE events B and C entered its DNA and then was passed on to its descendants, yet this occurred after the point in time where an earlier common ancestor had given rise both to that species, and to the lineage which later became pigs.
And this pattern (giving the *same* results) is repeated over and over and over again when various kinds of molecular evidence from DNA is examined in detail.
The molecular evidence for evolution and common descent is overwhelming. The only alternative is for creationists to deny the obvious and say, "well maybe God decided to set up all DNA in *only* ways that were consistent with an evolutionary result even though He'd have a lot more options open to him, even including parts which by every measure are useless and exactly mimic copy errors, ancient infections, stutters, and other garbage inherited from nonexistent shared ancestors"...
Humans have 23 pairs of chromosomes ---chimps and gorillas have 24 pairs. How many pairs of chromosomes did the "common ancestor" have? Was it 23 or 24 pairs? How do you "evolve" missing or added chromosomes ---that would happen all at one time.And:The common ancestor had 24 chromosomes.
If you look at the gene sequences, you'll find that Chromosome 2 in humans is pretty much just 2 shorter chimpanzee chromosomes pasted end-to-end, with perhaps a slight bit of lost overlap:
(H=Human, C=Chimpanzee, G=Gorilla, O=Orangutan)
Somewhere along the line, after humans split off from the other great apes, or during the split itself, there was an accidental fusion of two chromosomes, end-to-end. Where there used to be 24 chromosomes, now there were 23, but containing the same total genes, so other than a "repackaging", the DNA "instructions" remained the same.
If a chimpanzee gives birth to a creature with 23 chromosomes, that offspring isn't going to be a well-formed chimpanzee able to survive well.
It is if the same genes are present, which they would be in the case of a chromosome fusion.
Evolve would imply the genetic material changes little by little --not some big loss of two chromosomes at once but I don't see how they'd go away gene by gene.
Tacking two chromosomes together end-to-end is not a "big loss" of genes, and it really is a "little by little" change in the total genetic code. It's just been "regrouped" a bit. Instead of coming in 24 "packages", it's now contained in 23, but the contents are the same.
So how, you might ask, would the chromosomes from the first 23-chromosome "fused" individual match up with the 24 chromosomes from its mate when it tried to produce offspring? Very well, thanks for asking. The "top half" of the new extra-long Chromosome 2 would adhere to the original chromosome (call it "2p") from which it was formed, and likewise for the "bottom half" which would adhere to the other original shorter chromosome (call it "2q"). In the picture above, imagine the two chimp chromosomes sliding over to "match up" against the human chromosome. The chimp chromosomes would end up butting ends with each other, or slightly overlapping in a "kink", but chromosomes have overcome worse mismatches (just consider the XY pair in every human male -- the X and the Y chromosome are *very* different in shape, length, and structure, but they still pair up).
In fact, the "rubbing ends" of the matched-up chimp chromosomes, adhering to the double-long human-type chromosome, would be more likely to become fused together themselves.
For studies in which recent chromosome fusions have been discovered and found not to cause infertility, see:
Chromosomal heterozygosity and fertility in house mice (Mus musculus domesticus) from Northern Italy. Hauffe HC, Searle JB Department of Zoology, University of Oxford, Oxford OX1 3PS, United Kingdom. hauffe@novanet.itIn that last reference, the Przewalski horse, which has 33 chromosomes, and the domestic horse, with 32 chromosomes (due to a fusion), are able to mate and produce fertile offspring.An observed chromosome fusion: Hereditas 1998;129(2):177-80 A new centric fusion translocation in cattle: rob (13;19). Molteni L, De Giovanni-Macchi A, Succi G, Cremonesi F, Stacchezzini S, Di Meo GP, Iannuzzi L Institute of Animal Husbandry, Faculty of Agricultural Science, Milan, Italy.
J Reprod Fertil 1979 Nov;57(2):363-75 Cytogenetics and reproduction of sheep with multiple centric fusions (Robertsonian translocations). Bruere AN, Ellis PM
J Reprod Fertil Suppl 1975 Oct;(23):356-70 Cytogenetic studies of three equine hybrids. Chandley AC, Short RV, Allen WR.
Meanwhile, the question may be asked, how do we know that the human Chromosome 2 is actually the result of a chromsome fusion at/since a common ancestor, and not simply a matter of "different design"?
Well, if two chromsomes accidentally merged, there should be molecular remnants of the original chromosomal structures (while a chromosome designed from scratch would have no need for such leftover "train-wreck" pieces).
Ends of chromosomes have characteristic DNA base-pair sequences called "telomeres". And there are indeed remnants of telomeres at the point of presumed fusion on human Chromosome 2 (i.e., where the two ancestral ape chromosomes merged end-to-end). If I may crib from a web page:
Telomeres in humans have been shown to consist of head to tail repeats of the bases 5'TTAGGG running toward the end of the chromosome. Furthermore, there is a characteristic pattern of the base pairs in what is called the pre-telomeric region, the region just before the telomere. When the vicinity of chromosome 2 where the fusion is expected to occur (based on comparison to chimp chromosomes 2p and 2q) is examined, we see first sequences that are characteristic of the pre-telomeric region, then a section of telomeric sequences, and then another section of pre-telomeric sequences. Furthermore, in the telomeric section, it is observed that there is a point where instead of being arranged head to tail, the telomeric repeats suddenly reverse direction - becoming (CCCTAA)3' instead of 5'(TTAGGG), and the second pre-telomeric section is also the reverse of the first telomeric section. This pattern is precisely as predicted by a telomere to telomere fusion of the chimpanzee (ancestor) 2p and 2q chromosomes, and in precisely the expected location. Note that the CCCTAA sequence is the reversed complement of TTAGGG (C pairs with G, and T pairs with A).Another piece of evidence is that if human Chromosome 2 had formed by chromosome fusion in an ancestor instead of being designed "as is", it should have evidence of 2 centromeres (the "pinched waist" in the picture above -- chromosomes have centromeres to aid in cell division). A "designed" chromosome would need only 1 centromere. An accidentally "merged" chromosome would show evidence of the 2 centromeres from the two chromosomes it merged from (one from each). And indeed, as documented in (Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M, Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet 1992 May;89(2):247-9), the functional centromere found on human Chromosome 2 lines up with the centromere of the chimp 2p chromosome, while there are non-functional remnants of the chimp 2q centromere at the expected location on the human chromosome.As an aside, the next time some creationist claims that there is "no evidence" for common ancestry or evolution, keep in mind that the sort of detailed "detective story" discussed above is repeated literally COUNTLESS times in the ordinary pursuit of scientific research and examination of biological and other types of evidence. Common ancestry and evolution is confirmed in bit and little ways over and over and over again. It's not just something that a couple of whacky anti-religionists dream up out of thin air and promulgate for no reason, as the creationists would have you believe.
[The poster known as Mr. LLLICHY wrote:] Here is that Vitamin C dataSee also, for example (out of thousands):After discovering this same data on another thread along with more discussion than has appeared here (I've taken the liberty of pinging the participants of that discussion), I see what the "mystery" is supposed to be -- it's supposed be why did some sites have multiple mutations while (small) stretches of other sites had none? In other words, why do the mutations appear clustered?
(You know, it would really help if people explained their points and questions in more detail, instead of leaving people to guess what the poster was thinking...)
[LLLICHY wrote:] "U238" that decays thrice, pretty good trick when there is "U238" that does not decay at all in 50,000,000 years.
Actually, no site had mutations "thrice". Three different bases at a given site is only *two* mutations (one original base, plus two mutations from it to something else).
Here's the "mutation map" from the actual DNA data:
--1-12--1-1-1-1--------1112112--1---1-11-1--------1 ALL/nNo mutations ("-") in about half the sites, one mutation at several (17) sites, two mutations at three sites.The first thing to keep in mind that random processes tend to "cluster" more than people expect anyway. People expect "randomness" to "spread out" somewhat evenly, but instead it's usually more "clumped", for statistical reasons that would be a diversion to go into right now. So "that looks uneven" isn't always a good indication that something truly is non-random.
If you don't believe me on that, I wrote a program which made 23 mutations totally at random on a 51-site sequence, then repeated the process to see what different random outcomes would look like:
10 X$=STRING$(51,"-") 20 FOR I=1 TO 23 30 J%=INT(RND*51)+1 40 C$=MID$(X$,J%,1) 50 IF C$="-" THEN MID$(X$,J%,1)="1" ELSE MID$(X$,J%,1)=CHR$(ASC(C$)+1) 60 NEXT I 70 PRINT X$ 80 GOTO 10Yeah, it's BASIC, so sue me. Here's a typical screenful of the results:-21---1---2---111----2-----2-1121-------1---1--11-1 -1--1--21-11---1-1--1-1---1----1---21-11111---11--- 3-11---3-----1-----11-2-1---1--1----3--2---1--1---- ---1-1--22--1-1--2-2111--1-1111---1------1-------1- ---32----1-11-1-----1---2-231----1------1-----11--1 ----2---21--1---4----1-------------11-1--111-11-211 11--1-1---1-----1--1------1----3111--1----111-2-1-2 1112---1-3-1----1-1-----1-1------121--111-------1-1 -111121--1----1----1-1-1-1-11-2---1-1-------1-111-- -----------11-1---11-11--------21----12211--1---131 --1-211-1-1----21--11-1-2----1--1----11---11-----11 12---1-13------------2---21-21---11-1-1-1--2------- -----2-1---1-1----21--11-11-1---111-1--111-----2--1 -----1-----1-1-1-1---1-2----11-21-11--1-111---1-21- ---11--1-1-122-1-1-1--1-----2-1-1-1-------1-1---111 --2--11----2--1---12-2----1-1---1-1--1--12----1-1-1 -111-1-----1-1----------1-21111--1-2-11-11-1----11- 11-1--211-1221-----1--1-----11--1-2-1----------11-- -----1-12-11---2-1---11--1-2--1----11---111-1----11 11----1--12---12----1---31---1-11----2--1-11-1----- ---1--111-1--1-1-111----1-21----1-1-3---1------2--1 -2-11----1-1------1------2-1-1--111-111-1-1----1111 1--1--1-1---1-111111--2--1-1------112----2---11----Notice how oddly "clustered" most of them look, including one run which left a 13-site stretch "absolutely untouched", contrary to intuition (while having *4* mutations at a single site!)Frankly, I don't see anything in the real-life DNA mutation map which looks any different from these truly random runs. Random events tend to cluster more than people expect. That solves the "mystery" right there.
Also, there may be a selection factor -- the GLO gene is a *lot* bigger than this. One has to wonder if this small 51-bp section was presented just because it was the one that looked "least random". That would be a no-no, since one can always hand-select the most deviant subset out of larger sample in order to artificially skew the picture.
However, since there are some interesting evolutionary observations to be made, let's look at that DNA data again, slightly rearranged:
TAC CCC GTG GAG GTG CGC TTC ACT CGG GCG GAC GAC ATC CTG CTG AGC CCC PIG TAC CCC GTG GAG GTA CGC TTC ACT CGC GGG GAC GAC ATC CTG CTG AGC CCC BOS TAC CCC GTA GAG GTG CGC TTC ACC CGA GGC GAT GAC ATT CTG CTG AGC CCC RAT TAC CCC GTG GAG GTG CGC TTC ACC CGA GGT GAT GAC ATC CTG CTG AGC CCG MOUSE TAC CCT GTG GGG GTG CGC TTC ACC CGG GGG GAC GAC ATC CTG CTG AGC CCC GUIN PIG TAC CTG GTG GGG GTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC HUMAN TAC CTG GTG GGG CTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC CHIMPANZEE TAC CCG GTG GGG GTG CGC TTC ACC CAG AG* GAT GAC GTC CTA CTG AGC CCC ORANGUTAN TAA CCG GTG GGG GTG CGC TTC ACC CAA GG* GAT GAC ATC ATA CTG AGC CCC MACAQUEHere I've put spaces between codons, and clustered the closely-related species together: pig/cow as ungulates, rat/mouse for their obvious relationship, guinea pig right below them but separated because of the pseudogene nature of its GLO gene, then primates all in a group, with man's closest relative, the chimp, immediately below him, followed by the more distant orangutan, and the even more distant macaque. Also note that the top four have "working" GLO genes, and the bottom five have "broken" GLO pseudogenes.First, let's consider just the four species with working GLO genes. Evolution predicts that even over large periods of time, these genes will be "highly conserved", with natural selection weeding out mutations that could "break" the gene. Note that the mutations will still have occurred in individuals of the population, but natural selection will "discourage" that mutation from spreading into the general population.
And before we go any further, let's talk about the "universal genetic code". In all mammals (indeed, in almost all living organisms), each triplet of DNA sites cause a particular amino acid to be formed. The mapping of triplets (called "codons") to amino acids is as follows:
Second Position of Codon T C A G F
i
r
s
t
P
o
s
i
t
i
o
nT
TTT Phe [F] TTC Phe [F] TTA Leu [L] TTG Leu [L]
TCT Ser [S] TCC Ser [S] TCA Ser [S] TCG Ser [S]
TAT Tyr [Y] TAC Tyr [Y] TAA Ter [end] TAG Ter [end]
TGT Cys [C] TGC Cys [C] TGA Ter [end] TGG Trp [W]
T C A G T
h
i
r
d
P
o
s
i
t
i
o
nC
CTT Leu [L] CTC Leu [L] CTA Leu [L] CTG Leu [L]
CCT Pro [P] CCC Pro [P] CCA Pro [P] CCG Pro [P]
CAT His [H] CAC His [H] CAA Gln [Q] CAG Gln [Q]
CGT Arg [R] CGC Arg [R] CGA Arg [R] CGG Arg [R]
T C A G A
ATT Ile [I] ATC Ile [I] ATA Ile [I] ATG Met [M]
ACT Thr [T] ACC Thr [T] ACA Thr [T] ACG Thr [T]
AAT Asn [N] AAC Asn [N] AAA Lys [K] AAG Lys [K]
AGT Ser [S] AGC Ser [S] AGA Arg [R] AGG Arg [R]
T C A G G
GTT Val [V] GTC Val [V] GTA Val [V] GTG Val [V]
GCT Ala [A] GCC Ala [A] GCA Ala [A] GCG Ala [A]
GAT Asp [D] GAC Asp [D] GAA Glu [E] GAG Glu [E]
GGT Gly [G] GGC Gly [G] GGA Gly [G] GGG Gly [G]
T C A G (The above table imported from http://psyche.uthct.edu/shaun/SBlack/geneticd.html, which also has a nice introduction to the genetic code.)
Another version of the same table with nifty Java features and DNA database lookups can be found here.
The thing which is most relevant to the following discussion is the fact that most of the genetic codes are "redundant" -- more than one codon (triplet) encodes to exactly the same amino acid. This means that even in genes which are required for the organism, certain basepair mutations make absolutely no difference if the change is from one codon which maps into amino acid X to another codon which still maps into amino acid X. (This fact allows certain kinds of evolutionary "tracers" to be "read" from the DNA, as described here).
Now back to our DNA data. The redundancy in the genetic code means that some basepair sites will have more "degrees of freedom" than others (i.e., ways in which they can mutate without disrupting the gene's biological function in any way). Let's look at the four species with working GLO genes again:
TAC CCC GTG GAG GTG CGC TTC ACT CGG GCG GAC GAC ATC CTG CTG AGC CCC PIG TAC CCC GTG GAG GTA CGC TTC ACT CGC GGG GAC GAC ATC CTG CTG AGC CCC BOS TAC CCC GTA GAG GTG CGC TTC ACC CGA GGC GAT GAC ATT CTG CTG AGC CCC RAT TAC CCC GTG GAG GTG CGC TTC ACC CGA GGT GAT GAC ATC CTG CTG AGC CCG MOUSE T T T A T A T T T A T C C T T T T T T T T A A A A A C A A A A A G C G G G G G C C C --- --- --1 --- --1 --- --- --1 --2 -12 --1 --- --1 --- --- --- --1Under each site of the mouse DNA, I've listed the "alternative" bases which could be be substituted for the mouse base at that site WITHOUT ALTERING THE GENE'S FUNCTION (because of genetic code redundancy). And under that I show the "mutation map" of just those four species.Note that most of the "alternative" bases are in the third base of each codon, *and* that this is where all but one of the mutations have appeared. This is because these were the sites which were "free" to mutate in the way they did, because the mutation was genetically neutral. That doesn't mean that the first and second sites of each codon were immune from mutation, it's just that when mutations did occur at those sites, natural selection weeded them out quickly because they most likely "broke" the GLO gene for the individuals which received that mutuation. What we see above is the results after natural selection has already "filtered" the undesirable mutations and left the ones which "do no harm".
Additionally, the two sites which have mutated twice (i.e. have a "2" in the mutation map) are ones which had more "allowable" mutations. Also note that the sites which had the fewest allowable alternatives (only one alternate letter allowed) didn't have any mutations fix at those sites, which is unsurprising since a "safe" mutation would be less likely to occur there versus a site that "allowed" two or three alternatives.
All this is as predicted by evolutionary theory, you'll note.
It also explains the one anomoly of the original mutation map, which is that the mutation counts do tend to be higher at the third base of a codon.
However... What about the one exception? The pig DNA has had one mutation at a site which does not encode to exactly the same amino acid (which is the case for *all* the other ones). In the pig DNA, the GGG codon (mapping to Glycine) has changed to a GCG codon (mapping to Alanine). What's up with that? Well, one of two things. First and most likely, just as base values in codons have a built-in redundancy, so do the amino acids which make up the proteins which result from the DNA templates. In other words, certain amino acids can be substituted for other ones at some sites in given proteins without making any functional difference. (This "protein functional redundancy" also has implications for "evolutionary tracer" analysis, see here.) That may well be the case for Alanine versus Glycine in the GLO protein, but I'm not enough of a biochemist to be able to say. The other option is that it *does* make some difference in the function of the pig GLO protein, but not enough to "break" the vitamin-C synthesis (as proven by the fact that pigs *can* synthesize vitamin C). So one way or another, it's not a deal-breaker even though pig GLO will not be 100% identical to cow/mouse/rat GLO. It's yet another "allowable" mutation.
More interesting evolutionary observations: The number of mutational differences between pig/cow is 3, the number between mouse/rat is 4, and the difference between rat/cow is 7 -- all roughly as one would expect from the evolutionary relatedness of these animals (cows/pigs and rats/mice are each closer to each other than the rodents are to the ungulates).
Now let's take a close look at the guinea pig:
TAC CCT GTG GGG GTG CGC TTC ACC CGG GGG GAC GAC ATC CTG CTG AGC CCC GUIN PIG --- --1 --- -1- --- --- --- --- --1 --1 --1 --- --- --- --- --- ---The "mutation map" under the guinea pig DNA is compared to the mouse DNA. Fascinating: Note that four of the five mutations are in the third base of a codon, *and* are of the type "allowed" by the genetic code redundancy. This indicates strongly that most of the evolutionary divergence between guinea pigs and mice likely occurred while the guinea pig's ancestors still had a working GLO gene. This is the sort of prediction implied by the evolutionary theory which could be cross-checked by further research of various types, and if verified, would be yet further confirmation that evolutionary theory is likely correct. So far, evolutionary theory has been subjected to literally countless tests like this, large and small, and the vast majority of results have confirmed the evolutionary prediction. This track record is hard to explain if evolution is an invalid theory, as some assert...Finally, let's look over the primate DNA and mutation map (relative to each other):
TAC CTG GTG GGG GTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC HUMAN TAC CTG GTG GGG CTA CGC TTC ACC TGG AG* GAT GAC ATC CTA CTG AGC CCC CHIMPANZEE TAC CCG GTG GGG GTG CGC TTC ACC CAG AG* GAT GAC GTC CTA CTG AGC CCC ORANGUTAN TAA CCG GTG GGG GTG CGC TTC ACC CAA GG* GAT GAC ATC ATA CTG AGC CCC MACAQUE --1 -1- --- --- 1-1 --- --- --- 111 1-- --- --- 1-- 1-- --- --- ---Evolutionary theory predicts that because the GLO gene is "broken" in primates (i.e. is a pseudogene), mutations in it are highly likely to be neutral (i.e., make no difference, since it can't get much more broken), and thus mutations are just as likely to accumulate at any site as any other. Is that what we see? Yup. There's no obvious pattern to the mutations between primates in the above mutation map, and unlike the pig/cow/mouse/rat mutation map, the mutations aren't predominantly at the "safer" third base of a codon, nor of a type that would be "safe". In fact, one base has vanished entirely, but no biggie, the gene's already broken.Also, although primates share a more recent common ancestor than cows/pigs/mice/rats, note that they've already racked up almost as many relative mutations as the cow/pig/mouse/rat DNA. This too is just as evolutionary theory predicts, because many mutations in a functional gene (GLO in this case) will be "non-safe" and weeded out by natural selection, making for a slower mutation fixation rate overall than in a pseudogene (as GLO is in primates) where natural selection doesn't "care" about the vast majority of mutations since *most* are neutral. So pseudogenes accumulate mutations faster than functional genes (even though rate of mutation *occurence* in both are likely the same).
Finally, note that there are ZERO mutational differences between the human DNA and the chimpanzee DNA, our nearest living relative.
I also see some interesting implications in the DNA sequences concerning which specific mutation fixed during what branch of the common-descent evolutionary tree for all the species represented, but reconstructing that would not only take another couple hours, at least, but would be a major bear to code in HTML, since I'd have to draw trees with annotations on the nodes... Bleugh.
In any case, I hope I've clarified some of the methods by which biologists find countless confirmations of evolution in DNA data. This is just a "baby" example, and to be more statistically valid would have to be done over much vaster sections of DNA sequences, but my intent was to demonstrate some of the concepts.
And if such a small amount of DNA as this can make small confirmations of evolutionary predictions, imagine the amount of confirmation from billion-basepair DNA data from each species compared across thousands of species... The amount of confirmatory discoveries for evolution from DNA analysis has already been vast, and promises to only grow in the future. For an overview of some of the different lines of evidence being studied, see The Journal of Molecular Evolution -- abstracts of all articles, current and back issues, can be browsed free online.
Analysis of the human Alu Ye lineageMolecular and temporal characteristics of human retropseudogenes.
Serine hydroxymethyltransferase pseudogene, SHMT-ps1: a unique genetic marker of the order primates
Insertions and duplications of mtDNA in the nuclear genomes of Old World monkeys and hominoids
Fixation times of retroposons in the ribosomal DNA spacer of human and other primates
The emergence of new DNA repeats and the divergence of primates
Genetic diversity at class II DRB loci of the primate MHC
Structure and evolution of human and African ape rDNA pseudogenes
Evolution predicted that transitional forms once existed between dinosaurian forelimbs and bird wings. Creationists predicted that "half a wing" would be unworkable and useless. Guess whose predictions were found to be right?Reptile -> Mammal evolutionary transition:
The above is from 29+ Evidences for Macroevolution, which compiles several hundred transitional fossils, which is itself just a *SMALL* sampling of the ENORMOUS numbers of fine transitional sequences found in the fossil record and well known to anyone who has bothered to CRACK OPEN A BOOK -- or even do a websearch -- in the past 25 years or so... So what's the anti-evolutionists' excuse for remaining abysmally ignorant of such things, and repeatedly making the false claim that there are "no" transitional fossils, etc.?Example 2: reptile-mammals
We also have an exquisitely complete series of fossils for the reptile-mammal intermediates, ranging from the pelycosauria, therapsida, cynodonta, up to primitive mammalia (Carroll 1988, pp. 392-396; Futuyma 1998, pp. 146-151; Gould 1990; Kardong 2002, pp. 255-275). As mentioned above, the standard phylogenetic tree indicates that mammals gradually evolved from a reptile-like ancestor, and that transitional species must have existed which were morphologically intermediate between reptiles and mammals—even though none are found living today. However, there are significant morphological differences between modern reptiles and modern mammals. Bones, of course, are what fossilize most readily, and that is where we look for transitional species from the past. Osteologically, two major striking differences exist between reptiles and mammals: (1) reptiles have at least four bones in the lower jaw (e.g. the dentary, articular, angular, surangular, and coronoid), while mammals have only one (the dentary), and (2) reptiles have only one middle ear bone (the stapes), while mammals have three (the hammer, anvil, and stapes) (see Figure 1.4.1).
Early in the 20th century, developmental biologists discovered something that further complicates the picture. In the reptilian fetus, two developing bones from the head eventually form two bones in the reptilian lower jaw, the quadrate and the articular (see the Pelycosaur in Figure 1.4.1). Surprisingly, the corresponding developing bones in the mammalian fetus eventually form the anvil and hammer of the unique mammalian middle ear (also known more formally as the incus and malleus, respectively; see Figure 1.4.2) (Gilbert 1997, pp. 894-896). These facts strongly indicated that the hammer and anvil had evolved from these reptilian jawbones—that is, if common descent was in fact true. This result was so striking, and the required intermediates so outlandish, that many anatomists had extreme trouble imagining how transitional forms bridging these morphologies could have existed while retaining function. Young-earth creationist Duane Gish stated the problem this way:
"All mammals, living or fossil, have a single bone, the dentary, on each side of the lower jaw, and all mammals, living or fossil, have three auditory ossicles or ear bones, the malleus, incus and stapes. ... Every reptile, living or fossil, however, has at least four bones in the lower jaw and only one auditory ossicle, the stapes. ... There are no transitional fossil forms showing, for instance, three or two jawbones, or two ear bones. No one has explained yet, for that matter, how the transitional form would have managed to chew while his jaw was being unhinged and rearticulated, or how he would hear while dragging two of his jaw bones up into his ear." (Gish 1978, p. 80)
Gish was incorrect in stating that there were no transitional fossil forms, and he has been corrected on this gaff numerous times since he wrote these words. However, Gish's statements nicely delineate the morphological conundrum at hand. Let's review the required evolutionary conclusion. During their evolution, two mammalian middle ear bones (the hammer and anvil, aka malleus and incus) were derived from two reptilian jawbones. Thus there was a major evolutionary transition in which several reptilian jawbones (the quadrate, articular, and angular) were extensively reduced and modified gradually to form the modern mammalian middle ear. At the same time, the dentary bone, a part of the reptilian jaw, was expanded to form the major mammalian lower jawbone. During the course of this change, the bones that form the hinge joint of the jaw changed identity. Importantly, the reptilian jaw joint is formed at the intersection of the quadrate and articular whereas the mammalian jaw joint is formed at the intersection of the squamosal and dentary (see Figure 1.4.1).
How could hearing and jaw articulation be preserved during this transition? As clearly shown from the many transitional fossils that have been found (see Figure 1.4.3), the bones that transfer sound in the reptilian and mammalian ear were in contact with each other throughout the evolution of this transition. In reptiles, the stapes contacts the quadrate, which in turn contacts the articular. In mammals, the stapes contacts the incus, which in turn contacts the malleus (see Figure 1.4.2). Since the quadrate evolved into the incus, and the articular evolved into the malleus, these three bones were in constant contact during this impressive evolutionary change. Furthermore, a functional jaw joint was maintained by redundancy—several of the intermediate fossils have both a reptilian jaw joint (from the quadrate and articular) and a mammalian jaw joint (from the dentary and squamosal). Several late cynodonts and Morganucodon clearly have a double-jointed jaw. In this way, the reptilian-style jaw joint was freed to evolve a new specialized function in the middle ear. It is worthy of note that some modern species of snakes have a double-jointed jaw involving different bones, so such a mechanical arrangement is certainly possible and functional.
Since Figure 1.4.3 was made, several important intermediate fossils have been discovered that fit between Morganucodon and the earliest mammals. These new discoveries include a complete skull of Hadrocodium wui (Luo et al. 2001) and cranial and jaw material from Repenomamus and Gobiconodon (Wang et al. 2001). These new fossil finds clarify exactly when and how the malleus, incus, and angular completely detached from the lower jaw and became solely auditory ear ossicles.
Recall that Gish stated: "There are no transitional fossil forms showing, for instance, three or two jawbones, or two ear bones" (Gish 1978, p. 80). Gish simply does not understand how gradual transitions happen (something he should understand, obviously, if he intends to criticize evolutionary theory). These fossil intermediates illustrate why Gish's statement is a gross mischaracterization of how a transitional form should look. In several of the known intermediates, the bones have overlapping functions, and one bone can be called both an ear bone and a jaw bone; these bones serve two functions. Thus, there is no reason to expect transitional forms with intermediate numbers of jaw bones or ear bones. For example, in Morganucodon, the quadrate (anvil) and the articular (hammer) serve as mammalian-style ear bones and reptilian jaw bones simultaneously. In fact, even in modern reptiles the quadrate and articular serve to transmit sound to the stapes and the inner ear (see Figure 1.4.2). The relevant transition, then, is a process where the ear bones, initially located in the lower jaw, become specialized in function by eventually detaching from the lower jaw and moving closer to the inner ear.
Here's another look:
(The above is from The Fossil Record: Evolution or "Scientific Creation", which is yet ANOTHER source the anti-evolutionists are obviously completely ignorant of -- not that that stops them from spouting off falsehoods about the subject anyway...Mammal-Like Reptiles
As previously stated, a succession of transitional fossils exists that link reptiles (Class Reptilia) and mammals (Class Mammalia). These particular reptiles are classifie as Subclass Synapsida. Presently, this is the best example of th e transformation of one major higher taxon into another. The morphologic changes that took place are well documented by fossils, beginning with animals essentially 100% reptilian and resulting in animals essentially 100% mammalian. Therefore, I have chosen this as the example to summarize in more detail (Table 1, Fig. 1).
![[Fig. 1b]](http://www.gcssepm.org/images/fossil_b.gif)
Skulls and jaws of synapsid reptiles and mammals; left column side view of skull; center column top view of skull; right column side view of lower jaw. Hylonomus modified from Carroll (1964, Figs. 2,6; 1968, Figs. 10-2, 10-5; note that Hylonomus is a protorothyrod, not a synapsid). Archaeothyris modified from Reisz (1972, Fig. 2). Haptodus modified from Currie (1977, Figs, 1a, 1b; 1979, Figs. 5a, 5b). Sphenacodo n modified from Romer & Price (1940, Fig. 4f), Allin (1975, p. 3, Fig. 16);note: Dimetrodon substituted for top view; modified from Romer & Price, 1940, pl. 10. Biarmosuchus modified from Ivakhnenko et al. (1997, pl. 65, Figs. 1a, 1B, 2); Alin & Hopson (1992; Fig. 28.4c); Sigogneau & Tchudinov (1972, Figs. 1, 15). Eoarctops modified from Broom (1932, Fig. 35a); Boonstra (1969, Fig. 18). Pristerognathus modified from Broom (1932, Figs 17a, b,c); Boonstra (1963, Fig. 5d). Procynosuchus modified from Allin & Hopson (1992, Fig. 28.4e); Hopson (1987, Fig. 5c); Brink (1963, Fig. 10a); Kemp (1979, Fig. 1); Allin (1975, p. 3, Fig. 14). Thrinaxodon modified from Allin & Hopson (1992, Fig. 28.4f);Parrington (1946, Fig. 1); Allin (1975, p. 3, Fig. 13). Probainognathus modified from Allin & Hopson (1992, Fig. 28.4g); Romer (1970, Fig. 1); Allin (1975, p. 3, Fig. 12). Morga nucodon modified from Kermack, Mussett, & Rigney (1981, Figs. 95, 99a; 1973, Fig. 7a); Allin (1975, p. 3, Fig. 11). Asioryctes modified from Carroll (1988, Fig. 20-3b). Abbreviations: ag = angular; ar = articular; cp = coronoid process; d = dentary; f = lateral temporal fenestra; j = jugal; mm = attachment site for mammalian jaw muscles; o = eye socket; po = post orbital; q = quadrate; rl = reflected lamina; sq = squamosal; ty = tympanic.
TAXONOMY LATERAL TEMPORAL FENESTRA LOWER JAW DENTARY TEETH LOWER JAW: POST- DENTARY BONES MIDDLE EAR & JAW ARTICULATION M: Early Placental mammals
Asioryctes
Upper CretaceousMerged with eye socket; cheek arch bowed out laterally 100% of jaw length is the den- tary; condylar process in contact with squamosal Fully differentiated teeth; incisors, canines, premolars; one tooth replacement No post-dentary bones 3 middle ear bones (stapes, incus, malleus) + tympanic; squamosal-dentary jaw joint L: "Pantothere" mammals
Amphitherium
Middle/Upper JurassicX 100% of jaw length is the den- tary; condylar process contacts squamosal Fully differentiated teeth; incisors, canines, premolars; one tooth replacement Post-dentary bones migrated to middle ear Probably 3 middle ear bones (stapes, incus, malleus) + tympanic; squamosal-dentary jaw joint K: Morganucodontid mammals
Morganucodon Upper Triassic & Lower JurassicMerged with eye socket; cheeck arch bowed out laterally 100% of jaw length is the den- tary; condylar process expanded posteriorly to make contact with squamosal Fully differentiated teeth; incisors, canines, premolars; one tooth replacement 20% of jaw length; reflected lamina decreased to narrow ribbon-like horseshoe Stapes extends from inner ear capsule to quadrate; quadrate tiny; both quadrate-articular and squamosal-dentary jaw joints J: Chiniquodontid cynodonts
Probainognathus
Middle TriassicMuch larger than eye socket; 40- 45% of skull length; expanded posterioirly, medially, & laterally; midline of skull narrow sagittal crest; chek arch bowed out laterally 95% of jaw length is the dentary; large coronoid process expanded posteriorly; condylar process expanded posteriorly Large single canine; cheek teeth multicusped; tooth replacement reduced 20% of jaw length; angular notch widened ventrally; width of main part of angular decreased; reflec - ted lamina decreased to narrow ribbon-like horseshoe Stapes extends from inner ear capsule to quadrate; quadrate tiny; quadrate-articular joint I:Galesaurid cynodonts
Thrinaxodon
Lower TriassicMuch larger than eye socket; 40% of skull length; expanded pos- terioirly, medially, & laterally; midline of skull narrow sagittal crest; chek arch bowed out laterally 85% of jaw length is the dentary; large coronoid process expanded to top of eye socket and pos- teriorly; jaw muscles attached to most of coronoid process Large single canine; cheek teeth multicusped; tooth replacement reduced 25% of jaw length; angular notch widened ventrally; width of reflec- ted lamina decreased; width of main part of angular decreased Stapes extends from inner ear capsule to quadrate; quadrate small; quadrate-articular jaw joint H: Procynosuchid cynodonts
Procynosuchus
upper Upper PermianMuch larger than eye socket; 40% of skull length; expanded pos- terioirly, medially, & laterally; midline of skull narrow sagittal crest; chek arch bowed out laterally 75-80% of jaw length is the den- tary; coronoid process expanded to near top of eye socket and posteriorly; jaw muscles attached to dorsal part of coronoid process Large single canine; cheek teeth multicusped 30% of jaw length; angular notch widened ventrally; width of reflected lamina decreased Stapes extends from inner ear capsule to quadrate; quadrate small; quadrate-articular jaw joint G: Early Therocephalians
Pristerognathus
lower Upper PermianLarger than eye socket; expanded posteriorly and medially; 30% of skull length 75-80% of jaw length is the den- tary; posterior end of dentary expanded posteriorly and dorsally into narrow blade-like coronoid process; rises to middle of eye socket Large single canine; other teeth simple cones. 35% of jaw length; angular notch deepened into a cleft; reflected lamina large, broad, blade-like Stapes extends from inner ear capsule to quadrate; quadrate small; quadrate-articular jaw joint F: Early Gorgonopsians
Eoarctops
lower Upper PermianSlightly larger than eye socket; expanded posteriorly and medially (minimal); 20-25% of skull length 65-75% of jaw length is the den- tary; posterior end of dentary slightly expanded posteriorly and dorsally as incipient coronoid process Large single canine; other teeth simple cones. 40% of jaw length; angular notch deepened into a cleft; reflected lamina large, broad, blade-like Stapes extends from inner ear capsule to quadrate; quadrate- articular jaw joint E: Eotitanosuchians
Sphenacodon
Lower PermianSmall; slightly smaller than eye socket; slightly expanded posteriorly and medially 65-75% of jaw length is the den- tary; posterodorsal edge rises broadly but slightly above tooth row Large single canine; other teeth simple cones. 40% of jaw length; angular notch deepened into a cleft; reflected lamina large, broad, blade-like Stapes extends from inner ear capsule to quadrate; quadrate- articular jaw joint D: Late sphenacodonts
Sphenacodon
Upper PennsylvanianSmall; smaller than eye socket; confined to one side of skull 65% of jaw length is the dentary; posterodorsal edge rises broadly but slightly above the tooth row Enlarged incipient canines; other teeth simple cones 60% of jaw length; venntral edge of angular notched ("angular notch") offsetting a short pro- tusion (reflected lamina) Stapes extends from inner ear capsule to quadrate; quadrate large and plate-like; quadrate- articular jaw joint C: Early spenacodonts
Haptodus
Upper PennsylvanianTiny; smaller than eye socket; confined to one side of skull 65-75% of jaw length is the den- tary; posterodorsal edge rises broadly but slightly above tooth row Undifferentiated; slightly enlarged incipient canines just behind nares 70% of jaw length; ventral edge of angular with shallow indentation Stapes extends from inner ear capsule to quadrate; quadrate- articular jaw joint B: Early ophiacodonts
Archaothyris
upper Middle PennsylvanianTiny; smaller than eye socket; confined to one side of skull x Undifferentiated; slightly enlarged incipient canines just behind nares x Stapes extends from inner ear capsule to quadrate; quadrate- articular jaw joint A: Protorothyrids
Hylonomus
lower Middle PennsylvanianAbsent 65-75% of jaw length is the den- tary; posterodorsal edge rises broadly but slightly above tooth row Undifferentiated; slightly enlarged incipient canines just behind nares 70% of jaw length; ventral edge of angular continuous Stapes extends from inner ear capsule to quadrate; quadrate- articular jaw joint
Table 1: Morphology of synapsid reptiles and mammals (Note that Hylonomus is a protothyrid, not a synapsid). Data from references cited in text.
Modern reptiles and mammals are very distinctive, easily diagnosable, and do not intergrade. Reptiles are covered by scales, mammals by hair; reptiles are cold-blooded, mammals warm-blooded; reptiles do not suckle their young, mammals have mammary glands; reptiles have sprawling posture, mammals have upright posture. Most of these features are soft part anatomy or physiology that very rarely fossilize (although dinosaur skin impressions are known from Cretaceous sediments, and imprints of mammal hair are known from Eocene bats from Germany; Franzen, 1990). In the fossil record, we must look to skeletal features.
There are many skeletal features which allow us to distinguish the reptiles from the mammals (Carroll, 1988; Table 1, rows A, M). The single most important defining characteristic is the nature of the articulation of the lower jaw to the skull (Simpson, 1959). In reptiles, multiple bones comprise the lower jaw. A small bone at the posterior end of the lower jaw, the articular, articulates with the quadrate bone of the skull (Simpson, 1959; Carroll, 1988). In mammals, one large bone, the dentary, comprises the lower jaw. It articulates with the squamosal bone of the skull (Simpson, 1959; Carroll, 1988).
From comparative anatomy studies, it is certain that most of the bones of the reptiles and mammals are homologous (Crompton & Parker, 1978; Carroll, 1988). Of greatest importance, the middle ear bones of mammals (stapes, incus, malleus, and tympanic) are homologous with several of the skull and jaw bones of reptiles (stapes, quadrate, articular, and angular, respectively; Romer, 1956, p. 33-38, 1970a; Allin, 1975, 1986; Allin & Hopson, 1992; Crompton & Parker, 1978; Hopso n, 1987, 1994; Carroll, 1988). One group of reptiles, the synapsids (Subclass Synapsida), share with the mammals an additional homologous structure: the lateral temporal fenestra, which is an opening in the skull behind the eye socket at the triple junction between the squamosal, jugal , and post orbital bones (Broom, 1932; Frazetta, 1968; Kemp, 1982; Carroll, 1988). A band of bone composed of the jugal and the squamosal is adjacent to the lateral temporal fenestra (Broom, 1932; Kemp, 1982; Carroll, 1988). This is the cheek arch so characteristic of mammal skulls (Broom, 1932; Kemp, 1982; Carroll, 1988). Therefore, synapsids are commonly named the “mammal-like reptiles.”
The presence of diagnosable morphologic differences between reptiles (including the oldest reptiles and the oldest synapsids) and mammals distinguishes them as distinct taxa. This allows us to test evolution by looking for transitional forms between the two. Because many of the bones are homologous, we should find evidence illustrating how these bones were modified over time to become the new bones. Furthermore, these morphologic changes should happen in parallel and in geochronologic succession.
Synapsid reptiles inhabited Pangea from the Middle Pennsylvanian through the Early Jurassic (Kemp, 1982, 1985; Sloan, 1983; Carroll, 1988; Hopson, 1969, 1987, 1994; Hopson & Crompton, 1969; Hotton, et al., 1986; Crompton & Jenkins, 1973; Sidor & Hopson, 1998; Romer & Price, 1940; Broom, 1932; Boonstra, 1963, 1969, 1971; Tchudinov, 1983; Olson, 1944; Tatarinov, 1974; Vyushkov, 1955; Efremov, 1954). From the Early Permian through the Early Triassic, they were the largest and most abundant land animals (Sloan, 1983; Colbert, 1965). Though much less well known to the general public than dinosaurs, one of the “cereal box dinosaurs,” Dimetrodon (the sail-backed reptile), is a synapsid, not a dinosaur (Romer & Price, 1940; Carroll, 1988). The oldest mammals are Late Triassic (Kemp, 1982; Carroll, 1988). Below is a discussion of the geochronologic succession linking synapsids and mammals. The oldest reptiles (named protorothyrids; Carroll, 1964, 1988, p. 192-199) are from the lower Middle Pennsylvanian, and the oldest synapsids (Reisz, 1972) are from the upper Middle Pennsylvanian, both of Nova Scotia. Upper Pennsylvanian and Lower Permian forms are known primarily from the midcontinent and Permian Basin region of the United States (Romer & Price, 1940; Currie, 1977, 1979; Kemp, 1982; Sloan, 1983). The basal Upper Permian forms are known from Russia (Tchudinov, 1960, 1983; Efremov, 1954; Olson, 1962; Sigogneau & Tchudinov, 1972; Ivakhnenko et al., 1997). Most of the Upper Permian and Lower Triassic succession is known from southern Africa, especially the Great Karoo of South Africa (Broom, 1932; Boonstra, 1963, 1969, 1971; Hopson & Kitching, 1972; Kemp, 1982; Sloan, 1983). The Middle Triassic forms are from South America (Romer, 1969a, 1969b, 1970b, 1973; Romer & Lewis, 1973; Bonaparte & Barbarena, 1975), and the Upper Triassic and Lower Jurassic mammals are known from Eurasia (Kermack, Mussett, & Rigney, 1973, 1981; Kemp, 1982). Subsequent Mesozoic mammals are known from all over the world (Simpson, 1928; Lillegraven et al., 1979).
When placed in proper geochronologic succession, the synapsids naturally form a succession of taxa (genera and families) that progressively become more mammal-like and less reptile-like (Kemp, 1982, 1985; Sloan, 1983; Sidor & Hopson, 1998; Hopson, 1987, 1994). Morphologic changes, summarized in Table 1 and Figure 1, affect the entire skeletal anatomy of these animals, but are most clearly displayed in their skulls.
The lateral temporal fenestra increased in size from a tiny opening smaller than the eye socket to a giant opening occupying nearly half the length of the skull. Ultimately, it merged with the eye socket, thus producing the full development of the cheek arch so characteristic of mammals (Broom, 1932; Frazetta, 1968; Kemp, 1982; Sloan, 1983; Hopson, 1987, 1994; Carroll, 1988).
Successively, the relative proportion of the lower jaw comprised of the dentary bone (teeth-bearing bone) gradually increased until the entire lower jaw consisted of the dentary (Kemp, 1982; Sloan, 1983; Carroll, 1988; Hopson, 1987, 1994). In Pennsylvanian and Lower and basal Upper Permian synapsids, the postero-dorsal edge of the lower jaw rose broadly but only slightly above the level of the tooth row (Romer & Price, 1940; Currie, 1977, 1979; Ivakhnenko et al., 1997; Tchudinov, 1960, 1983; Efremov, 1954; Olson, 1962; Sigogneau & Tchudinov, 1972; Hopson, 1987, 1994). In succeeding forms, the posterior part of the dentary expanded dorsally and posteriorly as a blade-like process, and progressively became larger (Broom, 1932; Boonstra, 1963, 1969, 1971; Sigogneau, 1970; Brink, 1963; Kemp, 1979; Hopson, 1987, 1994), forming the coronoid process (Parrington, 1946; Fourie, 1974; Romer, 1969b, 1970b, 1973; Hopson, 1987, 1994) to which the mammalian-type jaw musculature is attached (Barghusen, 1968; Bramble, 1978; Crompton, 1972; Crompton & Parker, 1978; Kemp, 1982; Sloan, 1983; Carroll, 1988). Concomitantly, the post-dentary bones progressively reduced in size (Allin, 1975; Crompton, 1972; Crompton & Parker, 1978; Kemp, 1982; Sloan, 1983; Carroll, 1988; Hopson, 1987, 1994).
Beginning with the Upper Pennsylvanian sphenacodonts, a notch developed in the angular bone that offsets a projection, the reflected lamina (Allin, 1975; Allin & Hopson, 1992; Hopson, 1987, 1994; Romer & Price, 1940; Currie, 1977, 1979; Kemp, 1982; Sloan, 1983; Carroll, 1988). The reflected lamina first became a large blade-like flange (Allin, 1975; Allin & Hopson, 1992; Hopson, 1987, 1994; Ivakhnenko et al., 1997; Tchudinov, 1960, 1983; Efremov, 1954; Olson, 1962; Sigogneau & Tchudinov, 1972; Broom, 1932; Sigogneau, 1970; Boonstra, 1963, 1969, 1971), and then was progressively reduced to a delicate horseshoe-shaped bone (Allin, 1975; Allin & Hopson, 1992; Hopson, 1987, 1994; Brink, 1963; Parrington, 1946; Fourie, 1974; Romer, 1969b, 1970b, 1973; Kermack, Mussett, & Rigney, 1973, 1981; Kemp, 1979, 1982; Sloan, 1983; Carroll, 1988).
Simultaneously, the quadrate progressively decreased in size (Allin, 1975; Allin & Hopson, 1992; Hopson, 1987, 1994; Kemp, 1982; Sloan, 1983; Carroll, 1988). The articular did not decrease in size much, being small initially, but developed a downward-pointing prong (Allin, 1975; Allin & Hopson, 1992; Hopson, 1987, 1994; Kemp, 1982; Sloan, 1983; Carroll, 1988). In the synapsids, the lower jaw was hinged to the skull by the articular and quadrate bones (Crompton, 1972; Crompton & Parker, 1978; Allin, 1975; Allin & Hopson, 1992; Hopson, 1987, 1994). Thus they are classified as reptiles (Simpson, 1959; Kemp, 1982; Sloan, 1983; Carroll, 1988). As the quadrate and articular became smaller, they were relieved of their solid suture to the dentary and skull (Crompton, 1972; Allin, 1975, 1986; Allin & Hopson, 1992; Hopson, 1987, 1994; Crompton & Parker, 1978; Kemp, 1982; Sloan, 1983; Carroll, 1988). A projection of the dentary extended posteriorly and made contact with the squamosal. Morganucodon possessed the mammalian dentary-squamosal jaw joint adjacent to the reptilian articular-quadrate jaw joint (Kermack, Mussett, & Rigney, 1973, 1981; Carroll, 1988). It is classified as the first mammal, but it is a perfect intermediate. Now that a new jaw joint was established, the quadrate and articular were subsequently relieved of that function (Crompton, 1972; Allin, 1975, 1986; Allin & Hopson, 1992; Hopson, 1987, 1994; Crompton & Parker, 1978; Kemp, 1982; Sloan, 1983; Carroll, 1988). Ultimately, in Middle and Upper Jurassic mammals, the tiny quadrate, articular, and ring-like angular migrated as a unit to the middle ear where they joined the stapes and became the incus, malleus, and tympanic bones (Allin, 197 5, 1986; Allin & Hopson, 1992; Hopson, 1987, 1994; Kemp, 1982; Sloan, 1983; Carroll, 1988).
Progressively, the teeth became differentiated. The large canines developed first, followed by the development of multicusped cheek teeth, reduced tooth replacement (Osborn & Crompton, 1973; Crompton & Parker, 1978), and finally full y differentiated incisors, canines, premolars, and molars with one tooth replacement during life (Kemp, 1982; Hopson, 1994).
Many other morphologic changes are documented in the fossil record. These demonstrate the morphologic and geochronologic succession from sprawling limb posture to upright limb posture of mammals (Jenkins, 1971; Romer & Lewis, 197 3; Kemp, 1982; Carroll, 1988; Hopson, 1994). As Jenkins (1971, p. 210) stated, “In details of morphology and function, the cynodont post-cranial skeleton should be regarded as neither ‘reptilian’ nor ‘mammalian’ but as transitional between the two classes .” Other changes have been adequately summarized elsewhere (Kemp, 1982; Sloan, 1983; Carroll, 1988; Hopson, 1994). Obviously, fundamental physiologic changes must have taken place as well, many of which are not directly preserved in the fossil record, though some can be inferred from the skeletal anatomy (Findlay, 1968; Kemp, 1982; Sloan, 1983, Carroll, 1988; Hopson, 1994).
This is well documented in the fossil record by a massive volume of incontrovertible data that cannot be explained away. Such large-scale, progressive, continuous, gradual, and geochronologically successive morphologic change (Sidor & Hopson, 1998) is descent with modification, and provides compelling evidence for evolution on a grand scale.
When the details of various biological mechanisms are examined, coherent evolutionary histories are discovered for them, again consistent with the predictions of evolutionary theory. For example, here are discoveries about the origins of the vertebrate clotting mechanism -- all consistent with the predictions of evolution:Theropod dinosaur to bird evolutionary transition:
The cladogram for the evolution of flight looks like this:
(Note -- each name along the top is a known transitional fossil; and those aren't all that have been discovered.) Here's a more detailed look at the middle section:
Fossils discovered in the past ten years in China have answered most of the "which came first" questions about the evolution of birds from dinosaurs.
We now know that downy feathers came first, as seen in this fossil of Sinosauropteryx:
That's a close-up of downy plumage along the backbone. Here's a shot of an entire fossil
Sinosauropteryx was reptilian in every way, not counting the feathers. It had short forelimbs, and the feathers were all the same size. Presumably, the downy feathers evolved from scales driven by a need for bodily insulation.
Next came Protarchaeopteryx:
It had long arms, broad "hands", and long claws:
Apparently this species was driven by selection to develop more efficient limbs for grasping prey. One of the interesting things about this species is that the structure of the forelimb has been refined to be quite efficient at sweeping out quickly to grab prey, snap the hands together, then draw them back towards the body (mouth?). The specific structures in question are the semilunate carpal (a wrist bone), that moves with the hand in a broad, flat, 190 degree arc, heavy chest muscles, bones of the arm which link together with the wrist so as to force the grasping hands to spread out toward the prey during the forestroke and fold in on the prey during the upstroke. Not only is this a marvelously efficient prey-grabbing mechanism, but the same mechanism is at the root of the wing flight-stroke of modern birds. Evolution often ends up developing a structure to serve one need, then finds it suitable for adaptation to another. Here, a prey-grasping motion similar in concept to the strike of a praying mantis in a reptile becomes suitable for modifying into a flapping flight motion.
Additionally, the feathers on the hands and tail have elongated, becoming better suited for helping to sweep prey into the hands.
Next is Caudipteryx:
This species had hand and tail feathers even more developed than the previous species, and longer feathers, more like that of modern birds:
However, it is clear that this was still not a free-flying animal yet, because the forelimbs were too short and the feathers not long enough to support its weight, and the feathers were symmetrical (equal sized "fins" on each side of the central quill). It also had very reduced teeth compared to earlier specimens and a stubby beak:
But the elongation of the feathers indicates some aerodynamic purpose, presumably gliding after leaping (or falling) from trees which it had climbed with its clawed limbs, in the manner of a flying squirrel. Feathers which were developed "for" heat retention and then pressed into service to help scoop prey were now "found" to be useful for breaking falls or gliding to cover distance (or swooping down on prey?).
Next is Sinornithosaurus:
Similar to the preceding species, except that the pubis bone has now shifted to point to the back instead of the front, a key feature in modern birds (when compared to the forward-facing publis bone in reptiles). Here are some of the forearm feathers in detail:
Long feathers in detail:
Artists' reconstruction:
Next is Archaeopteryx:
The transition to flight is now well underway. Archaeopteryx has the reversed hallux (thumb) characteristic of modern birds, and fully developed feathers of the type used for flight (long, aligned with each other, and assymetrical indicating that the feathers have been refined to function aerodynamically). The feathers and limbs are easily long enough to support the weight of this species in flight. However, it lacks some structures which would make endurance flying more practical (such as a keeled sternum for efficient anchoring of the pectoral muscles which power the downstroke) and fused chest vertebrae. Archaeopteryx also retains a number of clearly reptilian features still, including a clawed "hand" emerging from the wings, small reptilian teeth, and a long bony tail. After the previous species' gliding abilities gave it an advantage, evolution would have strongly selected for more improvements in "flying" ability, pushing the species towards something more resembling sustained powered flight.
Next is Confuciusornis:
This species had a nearly modern flight apparatus. It also displays transitional traits between a reptilian grasping "hand" and a fully formed wing as in modern birds -- the outer two digits (the earlier species had three-fingered "hands") in Confuciusornis are still free, but the center digit has now formed flat, broad bones as seen in the wings of modern birds.
Additionally, the foot is now well on its way towards being a perching foot as in modern birds:
It also has a keeled sternum better suited for long flight, and a reduced number of vertebrae in the tail, on its way towards becoming the truncated tail of modern birds (which while prominent, is a small flap of muscle made to look large only because of the long feathers attached).
From this species it's only a small number of minor changes to finish the transition into the modern bird family.
(Hey, who said there are no transitional fossils? Oh, right, a lot of dishonest creationists. And there are a lot more than this, I've just posted some of the more significant milestones.)
There's been a very recent fossil find along this same lineage, too new for me to have found any online images to include in this article. And analysis is still underway to determine exactly where it fits into the above lineage. But it has well-formed feathers, which extend out from both the "arms" and the legs. Although it wasn't advanced enough to fully fly, the balanced feathering on the front and back would have made it ideally suited for gliding like a flying squirrel, and it may be another link between the stage where feathers had not yet been pressed into service as aerodynamic aids, and the time when they began to be used more and more to catch the air and developing towards a "forelimbs as wings" specialization.
So in short, to answer your question about how flight could have developed in birds, the progression is most likely some minor refinement on the following:
1. Scales modified into downy feathers for heat retention.
2. Downy feathers modified into "straight" feathers for better heat retention (modern birds still use their body "contour feathers" in this fashion).
3. Straight feathers modified into a "grasping basket" on the hands (with an accompanying increase in reach for the same purpose).
4. Long limbs with long feathers refined to better survive falls to the ground.
5. "Parachute" feathers refined for better control, leading to gliding.
6. Gliding refined into better controlled, longer gliding.
7. Long gliding refined into short powered "hops".
8. Short powered flight refined into longer powered flight.
9. Longer powered flight refined into long-distance flying.Note that in each stage, the current configuration has already set the stage for natural selection to "prefer" individuals which better meet the requirements of the next stage. Evolution most often works like this; by taking some pre-existing ability or structure, and finding a better use for it or a better way to make it perform its current use.
Following are a bunch of papers which confirm various predictions of evolutionary biology. (Note, a recent change at NCBI has broken a lot of these links, but you can still find those papers by Googling for their titles):Evolution of the vertebrate clotting mechanism
In detail, discuss why gradual evolution of blood clotting with 10 protein feedback loops all working at once is actually quite feasible evolutionarily speaking.
Well, okay, since you insist... Check out The Evolution of Vertebrate Blood Clotting, or The evolution of vertebrate blood coagulation as viewed from a comparison of puffer fish and sea squirt genomes. Excerpt from the latter paper:
It is thought that 50–100 million years separate the appearances of urochordates (which include the sea squirt) and vertebrates. During that time the machinery for thrombin-catalyzed fibrin formation had to be concocted by gene duplication and the shuffling about of key modular domains. The relative times of duplicative events can be estimated by various means, the most obvious being the presence or absence of a gene in earlier diverging organisms, although it must be kept in mind that lineages may lose genes. Another way to gauge events is from the relative positions of various gene products on phylogenetic trees, earlier branching implying earlier appearance. In this regard, (pro)thrombin invariably appears lower on the phylogenetic trees than do the other vitamin K-dependent factors (Fig. 2).Also, Evolution of enzyme cascades from embryonic development to blood coagulation:The order of events can also be inferred by considering the most parsimonious route to assembling the various clusters of peripheral domains. Nine of the proteases under discussion can be accounted for by six domain-swapping events (Fig. 5). Indeed, the presence of a multiple-kringle protease in the sea squirt genome provides a reasonable model for a step-by-step parallel evolution of the clotting and lysis systems. It should be noted that a serine protease with only one kringle has been found in the ascidian Herdmania momus (36). Although numerous scenarios have been offered in the past about how modular exchange was involved in generating these schemes (refs. 4, 12, and 37–41, inter alia), the new genomic data now provide a realistic set of starting materials.
Recent delineation of the serine protease cascade controlling dorsal-ventral patterning during Drosophila embryogenesis allows this cascade to be compared with those controlling clotting and complement in vertebrates and invertebrates. The identification of discrete markers of serine protease evolution has made it possible to reconstruct the probable chronology of enzyme evolution and to gain new insights into functional linkages among the cascades. Here, it is proposed that a single ancestral developmental/immunity cascade gave rise to the protostome and deuterostome developmental, clotting and complement cascades. Extensive similarities suggest that these cascades were built by adding enzymes from the bottom of the cascade up and from similar macromolecular building blocks.That was the abstract. An excerpt from the text:The downstream protease of the vertebrate clotting cascade (Fig. 1d), thrombin, belongs to the same lineage as complement factors C1r and C1s. The upstream and middle proteases of the clotting cascade (factors VII, IX and X) belong to the most modern lineage, that of horseshoe crab clotting factor C. Therefore, the lineage of thrombin is parental to that of the upstream and middle proteases of the clotting cascade (Table 1) and distinguishes it from the other vitamin-K-dependent clotting proteases (factors VII, IX and X, and protein C). This conclusion agrees with sequence and species comparisons implying that thrombin was the ancestral blood-clotting protein [11]. It also suggests that vertebrate blood clotting emerged as a by-product of innate immunity, because the entire functional core of vertebrate clotting shares ancestry with complement proteases.And if that's not enough, you could check these out:Banyai, L., Varadi, A. and Patthy, L. (1983). “Common evolutionary origin of the fibrin-binding structures of fibronectin and tissue-type plasminogen activator.” FEBS Letters, 163(1): 37-41.
Bazan, J. F. (1990). “Structural design and molecular evolution of a cytokine receptor superfamily.” Proceedings of the National Academy of Sciences of the United States of America, 87(18): 6934-6938.
Blake, C. C. F., Harlos, K. and Holland, S. K. (1987). “Exon and Domain Evolution in the Proenzymes of Blood Coagulation and Fibrinolysis.” Cold Spring Harbor Symposia on Quantitative Biology: The Evolution of Catalytic Function, LII: 925-932.
Fornace AJ Jr, Cummings DE, Comeau CM, Kant JA, Crabtree GR. “The Structure of the human gamma-fibrinogen gene. Alternate mRNA splicing near the 3' end of the gene produces gamma A and gamma B forms of gamma-fibrinogen.” J Biol Chem. 1984 Oct 25;259(20):12826-30.
Crabtree, G. R., Comeau, C. M., Fowlkes, D. M., Fornace, A. J., Jr., Malley, J. D. and Kant, J. A. (1985). “Evolution and structure of the fibrinogen genes: Random insertion of introns or selective loss?” Journal of Molecular Biology, 185(1): 1-20.
Di Cera, E., Dang, Q. D. and Ayala, Y. M. (1997). “Molecular mechanisms of thrombin function.” Cell Mol Life Sci, 53(9): 701-730.
Doolittle, R. F. (1985). “More homologies among the vertebrate plasma proteins.” Biosci Rep, 5(10-11): 877-884.
Doolittle, R. F. (1990). “The Structure and Evolution of Vertebrate Fibrinogen A Comparison of the Lamprey and Mammalian Proteins,” in ADVANCES IN EXPERIMENTAL MEDICINE AND BIOLOGY: FIBRINOGEN, THROMBOSIS, COAGULATION, AND FIBRINOLYSIS. C. Y. Liu and Chien, S. New York, Plenum Press. 281.
Doolittle, R. F. (1992). “A detailed consideration of a principal domain of vertebrate fibrinogen and its relatives.” Protein Science, 1(12): 1563-1577.
Doolittle, R. F. (1992). “Early Evolution of the Vertebrate Fibrinogen Molecule.” Biophysical Journal, 61(2 PART 2): A410.
Doolittle, R. F. (1992). “Stein and Moore Award address. Reconstructing history with amino acid sequences.” Protein Science, 1(2): 191-200.
Doolittle, R. F. (1993). “The Evolution of Vertebrate Blood Coagulation - a Case of Yin and Yang.” Thrombosis and Haemostasis, V70(N1): 24-28.
Doolittle, R. F. and Feng, D. F. (1987). “Reconstructing the Evolution of Vertebrate Blood Coagulation from a Consideration of the Amino Acid Sequences of Clotting Proteins.” Cold Spring Harbor Symposia on Quantitative Biology: The Evolution of Catalytic Function, LII: 869-874.
Doolittle, R. F., G., Spraggon and J., Everse S. (1997). “Evolution of vertebrate fibrin formation and the process of its dissolution.” Ciba Found Symp, 212: 4-17; discussion 17-23.
Doolittle, R. F. and Riley, M. (1990). “The amino-terminal sequence of lobster fibrinogen reveals common ancestry with vitellogenins.” Biochemical and Biophysical Research Communications, 167(1): 16-19.
Edgington, T. S., Curtiss, L. K. and Plow, E. F. (1985). “A linkage between the hemostatic and immune systems embodied in the fibrinolytic release of lymphocyte suppressive peptides.” Journal of Immunology, 134(1): 471-477.
Ghidalia, W., Vendrely, R., Montmory, C., Coirault, Y., Samama, M., Lucet, B., Bellay, A. M. and Vergoz, D. (1989). “Overall study of the in vitro plasma clotting system in an invertebrate, Liocarcinus puber (Crustacea Decapoda): Considerations on the structure of the Crustacea plasma fibrinogen in relation to evolution.” Journal of Invertebrate Pathology, 53(2): 197-205.
Hervio, L. S., Coombs, G. S., Bergstrom, R. C., Trivedi, K., Corey, D. R. and Madison, E. L. (2000). “Negative selectivity and the evolution of protease cascades: the specificity of plasmin for peptide and protein substrates.” Chemistry & Biology, V7(N6): 443-452.
Hewett-Emmett, D., Czelusniak, J. and Goodman, M. (1981). “The evolutionary relationship of the enzymes involved in blood coagulation and hemostasis.” Annals of the New York Academy of Sciences, 370(20): 511-527.
Holland, S. K., Harlos, K. and Blake, C. C. F. (1987). “Deriving the generic structure of the fibronectin type II domain from the prothrombin Kringle 1 crystal structure.” EMBO (European Molecular Biology Organization) Journal, 6(7): 1875-1880.
Jordan, R. E. (1983). “Antithrombin in vertebrate species: conservation of the heparin-dependent anticoagulant mechanism.” Archives of Biochemistry and Biophysics, 227(2): 587-595.
Kant, J. A., Fornace, A. J., Jr., Saxe, D., Simon, M. J., McBride, O. W. and Crabtree, G. R. (1985). “Evolution and organization of the fibrinogen locus on chromosome 4: Gene duplication accompanied by transposition and inversion.” Proceedings of the National Academy of Sciences of the United States of America, 82(8): 2344-2348.
Kornblihtt, A. R., Pesce, C. G., Alonso, C. R., Cramer, P., Srebrow, A., Werbajh, S. and Muro, A. F. (1996). “The fibronectin gene as a model for splicing and transcription studies.” FASEB Journal, 10(2): 248-257.
Laki, K. (1972). “Our ancient heritage in blood clotting and some of its consequences.” Annals of the New York Academy of Sciences, 202(4): 297-307.
Neurath, H. (1984). “Evolution of proteolytic enzymes.” Science, 224(4647): 350-357.
Neurath, H. (1986). “The Versatility of Proteolytic Enzymes.” Journal of Cellular Biochemistry, 32(1): 35-50.
Oldberg, A. and Ruoslahti, E. (1986). “Evolution of the fibronectin gene: Exon structure of cell attachment domain.” Journal of Biological Chemistry, 261(5): 2113-2116.
Opal, S. M. (2000). “Phylogenetic and functional relationships between coagulation and the innate immune response.” Critical Care Medicine, V28(N9 SUPPS): S77-S80.
Pan, Y. and Doolittle, R. F. (1991). “Distribution of Introns in Lamprey Fibrinogen Genes.” Journal of Cellular Biochemistry Supplement(15 PART D): 75.
Pan, Y. and Doolittle, R. F. (1992). “cDNA sequence of a second fibrinogen alpha chain in lamprey: an archetypal version alignable with full-length beta and gamma chains.” Proceedings of the National Academy of Sciences of the United States of America, 89(6): 2066-2070.
Patthy, L. (1985). “Evolution of the Proteases of Blood Coagulation and Fibrinolysis by Assembly from Modules.” Cell, 41(3): 657-664.
Patthy, L. (1990). “Evolution of blood coagulation and fibrinolysis.” Blood Coagulation and Fibrinolysis, 1(2): 153-166.
Patthy, L. (1990). “Evolutionary Assembly of Blood Coagulation Proteins.” Seminars in Thrombosis and Hemostasis, 16(3): 245-259.
Patthy, L. (1999). “Genome evolution and the evolution of exon-shuffling—a review.” Gene, 238(1): 103-114.
Roberts, Lewis R., Nichols, Lanita A. and Holland, Lene J. (1995). “CDNA and amino-acid sequences and organization of the gene encoding the B-beta subunit of fibrinogen from Xenopus laevis.” Gene (Amsterdam), 160(2): 223-228.
Sosnoski, D. M., Emanuel, B. S., Hawkins, A. L., Van Tuinen, P., Ledbetter, D. H., Nussbaum, R. L., Kaos, F. T., Schwartz, E., Phillips, D. and et al. (1988). “Chromosomal localization of the genes for the vitronectin and fibronectin receptors .alpha. subunits and for platelet glycoproteins IIb and IIIa.” Journal of Clinical Investigation, 81(6): 1993-1998.
Wang, Y. Z., Patterson, J., Gray, J. E., Yu, C., Cottrell, B. A., Shimizu, A., Graham, D., Riley, M. and Doolittle, R. F. (1989). “Complete sequence of the lamprey fibrinogen .alpha. chain.” Biochemistry, 28(25): 9801-9806.
Xu, X. and Doolittle, R. F. (1990). “Presence of a vertebrate fibrinogen-like sequence in an echinoderm.” Proceedings of the National Academy of Sciences of the United States of America, 87(6): 2097-2101.
Zhang, Y. L., Hervio, L., Strandberg, L. and Madison, E. L. (1999). “Distinct contributions of residue 192 to the specificity of coagulation and fibrinolytic serine proteases.” Journal of Biological Chemistry, V274(N11): 7153-7156.
Zimmermann, E. (1983). “[The evolution of the coagulation system from primitive defense mechanisms].” Behring Institute Mitteilungen, 82(73): 1-12.
The 2.0-Å crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems
Davidson CJ, Tuddenham EG, McVey JH. 450 million years of hemostasis J Thromb Haemost. 2003 Jul;1(7):1487-94.
The Evolution of Improved Fitness by random mutation plus selection
Ancient Jumping DNA May Have Evolved Into Key Component Of Human Immune System
Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system
New insights into V(D)J recombination and its role in the evolution of the immune system
Evolution and developmental regulation of the major histocompatibility complex
Evolution of the IL-6/class IB cytokine receptor family in the immune and nervous systems
Development of an immune system
The ancestor of the adaptive immune system was the CAM system for organogenesis
The evolutionary origins of immunoglobulins and T-cell receptors: possibilities and probabilities
Evolutionary perspectives on amyloid and inflammatory features of Alzheimer disease
Organization of the human RH50A gene (RHAG) and evolution of base composition of the RH gene family.
Molecular evolution of the vertebrate immune system.
Morphostasis: an evolving perspective.
Rapid evolution of immunoglobulin superfamily C2 domains expressed in immune system cells.
Evolutionary assembly of blood coagulation proteins
Exon and domain evolution in the proenzymes of blood coagulation and fibrinolysis
Evolution of proteolytic enzymes
Evolution of vertebrate fibrin formation and the process of its dissolution.
Common Parasite Overturns Traditional Beliefs About The Evolution And Role Of Hemoglobin
Scientists Discover How Bacteria Protect Themselves Against Immune System
Reduction of two functional gamma-globin genes to one: an evolutionary trend in New World monkeys
Evolutionary history of introns in a multidomain globin gene
Hemoglobin A2: origin, evolution, and aftermath
Early evolution of microtubules and undulipodia
Flagellar beat patterns and their possible evolution
The evolutionary origin and phylogeny of eukaryote flagella
Dynein family of motor proteins: present status and future questions
Origins of the nucleate organisms
The evolutionary origin and phylogeny of microtubules, mitotic spindles and eukaryote flagella
The evolution of cellular movement in eukaryotes: the role of microfilaments and microtubules
Kinesin Motor Phylogenetic Tree
Evolution of a dynamic cytoskeleton
Isolation, characterization and evolution of nine pufferfish (Fugu rubripes) actin genes
Evolution of chordate actin genes: evidence from genomic organization and amino acid sequences
Co-evolution of ligand-receptor pairs in the vasopressin/oxytocin superfamily of bioactive peptides
The evolution of the synapses in the vertebrate central nervous system
Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria.
The evolution of metabolic cycles
Evolution of the first metabolic cycles
Speculations on the origin and evolution of metabolism
The Molecular Anatomy of an Ancient Adaptive Event
Biochemical pathways in prokaryotes can be traced backward through evolutionary time
Enzyme specialization during the evolution of amino acid biosynthetic pathways
Enzyme recruitment in evolution of new function
Bioenergetics: the evolution of molecular mechanisms and the development of bioenergetic concepts
Theoretical approaches to the evolutionary optimization of glycolysis--chemical analysis
The evolution of kinetoplastid glycosomes
Stepwise molecular evolution of bacterial photosynthetic energy conversion
Evolution of photosynthetic reaction centers and light harvesting chlorophyll proteins
Evolution of photosynthetic reaction centers
Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins
Evolution of the control of pigment and plastid development in photosynthetic organisms
Chemical evolution of photosynthesis
Molecular evolution of ruminant lysozymes
Evolution of stomach lysozyme: the pig lysozyme gene
Molecular basis for tetrachromatic color vision
Molecular evolution of the Rh3 gene in Drosophila
The evolution of rhodopsins and neurotransmitter receptors
Optimization, constraint, and history in the evolution of eyes
A pessimistic estimate of the time required for an eye to evolve
The eye of the blind mole rat (Spalax ehrenbergi): regressive evolution at the molecular level
Programming the Drosophila embryo
Evolution of chordate hox gene clusters
Hox genes in brachiopods and priapulids and protostome evolution.
Radical evolutionary change possible in a few generations
Evolution Re-Sculpted Animal Limbs By Genetic Switches Once Thought Too Drastic For Survival
The origin and evolution of animal appendages
Hox genes in evolution: protein surfaces and paralog groups
Evolution of the insect body plan as revealed by the Sex combs reduced expression pattern
Sea urchin Hox genes: insights into the ancestral Hox cluster
Theoretical approaches to the analysis of homeobox gene evolution
Teleost HoxD and HoxA genes: comparison with tetrapods and functional evolution of the HOXD complex
Evolutionary origin of insect wings from ancestral gills
Tracing backbone evolution through a tunicate's lost tail
The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster.
Gene duplications in evolution of archaeal family B DNA polymerases
Adaptive amino acid replacements accompanied by domain fusion in reverse transcriptase
Molecular evolution of genes encoding ribonucleases in ruminant species
Studies on the sites expressing evolutionary changes in the structure of eukaryotic 5S ribosomal RNA
Evolution of a Transfer RNA Gene Through a Point Mutation in the Anticodon
Universally conserved translation initiation factors
Genetic code in evolution: switching species-specific aminoacylation with a peptide transplant
Evolution of transcriptional regulatory elements within the promoter of a mammalian gene.
Codon reassignment and amino acid composition in hemichordate mitochondria.
Determining divergence times of the major kingdoms of living organisms with a protein clock
The multiplicity of domains in proteins
Characterization, primary structure, and evolution of lamprey plasma albumin
The origins and evolution of eukaryotic proteins
Evolution of vertebrate fibrin formation and the process of its dissolution.
Vastly Different Virus Families May Be Related
Selective sweep of a newly evolved sperm-specific gene in Drosophila
Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions
Molecular evolution of the histidine biosynthetic pathway
Accelerated evolution in inhibitor domains of porcine elafin family members
Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod
Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish
Evolution of an antifreeze glycoprotein
A model for the evolution of the plastid sec apparatus inferred from secY gene phylogeny
The evolutionary history of the amylase multigene family in Drosophila pseudoobscura
The evolution of an allosteric site in phosphorylase
Molecular evolution of fish neurohypophysial hormones: neutral and selective evolutionary mechanisms
Pseudogenes in ribonuclease evolution: a source of new biomacromolecular function?
Evolutionary relationships of the carbamoylphosphate synthetase genes
The molecular evolution of the small heat-shock proteins in plants
Evolutionary history of the 11p15 human mucin gene family.
Molecular evolution of the aldo-keto reductase gene superfamily.
A Classification of Possible Routes of Darwinian Evolution
Generation of evolutionary novelty by functional shift
Mobile DNA Sequences Could Be The Cause Of Chromosomal Mutations During The Evolution Of Species
Minor Shuffle Makes Protein Fold
Genetic Stowaways May Contribute To Evolutionary Change
Evolutionary Molecular Mechanism In Mammals Found
Cases of ancient mobile element DNA insertions that now affect gene regulation
Punctuated evolution caused by selection of rare beneficial mutations
The origin of programmed cell death
The origin and early development of biological catalysts
DNA secondary structures and the evolution of hypervariable tandem arrays
Episodic adaptive evolution of primate lysozymes
Genome plasticity as a paradigm of eubacteria evolution
Evolutionary motif and its biological and structural significance
Exon shuffling and other ways of module exchange
New Drosophila introns originate by duplication.
Evolution and the structural domains of proteins
The role of constrained self-organization in genome structural evolution
The coevolution of gene family trees
The evolution of metabolic cycles
The emergence of major cellular processes in evolution
A hardware interpretation of the evolution of the genetic code
Speculations on the origin and evolution of metabolism
Probabilistic reconstruction of ancestral protein sequences
The contribution of slippage-like processes to genome evolution
Molecular evolution in bacteria
The structural basis of molecular adaptation.
Mitochondrial DNA: molecular fossils in the nucleus
Cases of ancient mobile element DNA insertions that now affect gene regulation
Tiggers and DNA transposon fossils in the human genome
The eye of the blind mole rat (Spalax ehrenbergi): regressive evolution at the molecular level
Tiggers and DNA transposon fossils in the human genome
Gene competition and the possible evolutionary role of tumours
New Scientist Planet Science: Replaying life
Molecular evolution of an arsenate detoxification pathway by DNA shuffling
UB Researcher Developing Method That Employs Evolution To Develop New Drug Leads
Exploring the functional robustness of an enzyme by in vitro evolution
Evolutionary algorithms in computer-aided molecular design
Evolution of Enzymes for the Metabolism of New Chemical Inputs into the Environment
Evolution of Amino Acid Metabolism Inferred through Cladistic Analysis
Integrating the Universal Metabolism into a Phylogenetic Analysis
Serial segmental duplications during primate evolution result in complex human genome architecture
Phylogeny determined by protein domain content
Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily
Molecular archaeology of the Escherichia coli genome
Comparative Genomics of the Eukaryotes
Asymmetric Sequence Divergence of Duplicate Genes
The Genetic Core of the Universal Ancestor
Evolutionary History of Chromosome 20
Reconstructing large regions of an ancestral mammalian genome in silico
Occurrence and Consequences of Coding Sequence Insertions and Deletions in Mammalian Genomes
The Origin of Human Chromosome 1 and Its Homologs in Placental Mammals
On the RNA World: Evidence in Favor of an Early Ribonucleopeptide World
Inhibition of Ribozymes by Deoxyribonucleotides and the Origin of DNA
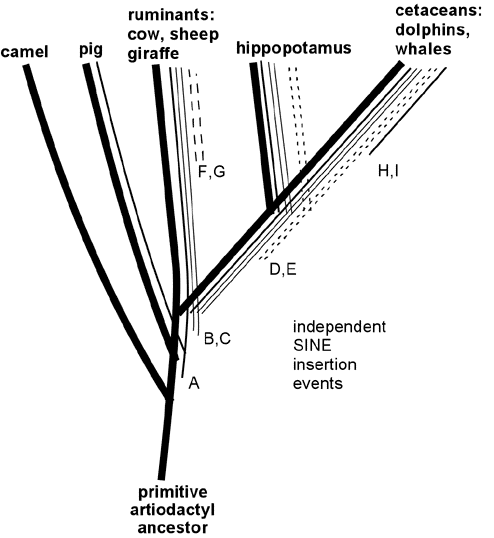

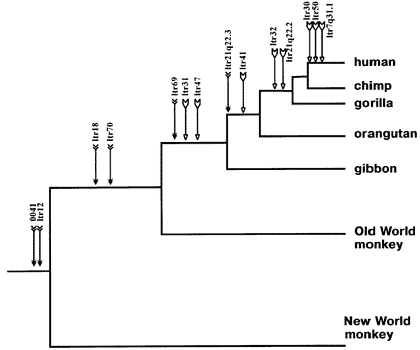

![[Figure1.4.1 (cartoon of vertebrate jaws)]](http://www.talkorigins.org/faqs/comdesc/images/jaws2.gif)
![[Figure1.4.2a (cartoon of vertebrate ears)]](http://www.talkorigins.org/faqs/comdesc/images/reptile_ear.gif)
![[Figure1.4.2b (cartoon of vertebrate ears)]](http://www.talkorigins.org/faqs/comdesc/images/human_ear.gif)
![[Figure1.4.3 (cartoon of vertebrate jaws)]](http://www.talkorigins.org/faqs/comdesc/images/jaws1.gif)
![[Fig. 1a]](http://www.gcssepm.org/images/fossil_a.gif)
















